Research Highlights
THE DOUBLE-HELIX: NOT JUST FOR DNA
1.0 Å crystal structure of parallel double-stranded poly(A) RNA published...
Aug. 12, 2013
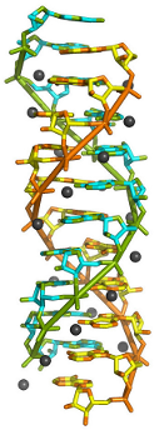
RNA (ribonucleic acid) sequences, although usually single-stranded, can sometimes form double-helical structures. Long RNA sequences having repeating adenine residues, poly(rA), are present on messenger RNA (mRNA), which is transcribed from DNA as a step toward the production of proteins. Poly(rA) RNA was predicted to have the ability to form a double-helix in 1961 based on fibre diffraction experiments. Its detailed structure has only been confirmed by X-ray crystallography recently. Researchers at McGill University have crystallized (rA)11 RNA sequences and combined data collected at the Canadian Light Source and the Cornell High Energy Synchrotron Source to obtain a very detailed 1.0 Å resolution crystal structure of this double-helix. The structure, obtained at physiological pH, shows a parallel double-helix. Ammonium ions stabilize the structure by binding to RNA phosphate groups and adenine N1 atoms, while N7 positions are engaged in hydrogen bonding. Contrary to antiparallel DNA, the poly(rA) double helix shows no major or minor grooves, but rather grooves of equal size. The extent of poly(rA) RNA double-helix formation in mRNA and in other systems remains to be discovered. Researchers believe the structure may be physiologically important, especially under conditions where there is a high local concentration of poly(rA). This can happen, for instance, under conditions where cells are stressed and mRNA become concentrated in RNA granules within cells. PDB ID: 4jrd.
Angew. Chem. Int. Ed. Engl., 52, 10370-10373